More Information
Submitted: December 23, 2022 | Approved: December 30, 2022 | Published: December 31, 2022
How to cite this article: Tonini D, Wu K, Saha R, Wang JP. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022; 6: 019-029.
DOI: 10.29328/journal.abse.1001018
Copyright License: © 2022 Tonini D, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Neuron activity; Neuronal networks; Magnetic recording; Magnetic field sensor; Computational neurobiology
Feasibility study of magnetic sensing for detecting single-neuron action potentials
Denis Tonini, Kai Wu, Renata Saha and Jian-Ping Wang*
Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN 55455, United States
*Address for Correspondence: Jian-Ping Wang, Department of Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN 55455, United States, Email: [email protected]
Understanding the magnitude of the local magnetic fields generated by neurons is critical to assessing the feasibility of novel magnetic field sensors to record in vivo neuronal activities at cellular resolution. However, the strength of the magnetic fields induced by individual neurons and neuronal networks has not been systematically studied. This step is critical for evaluating and benchmarking the ability of different magnetic field sensors to record neuronal activities with far better spatial and temporal resolution. Herein, FEM exemplary models and open-source computational libraries are used to calculate the magnetic fields generated by individual neurons and neuronal networks at micrometer distances. Our theoretical results show that the magnetic field generated by a single-neuron action potential can be detected by ultra-high sensitivity sub-pT magnetic field sensors, which opens the door to future in vivo decoding of neuronal activities through custom neural networks. We anticipate that the identification of single-neuron signals with high-sensitivity magnetic devices will allow the interface of nanoscale devices to interpret biological signals supported by machine-learning techniques capable of monitoring and predicting the localized activities underlying brain computations.
Nowadays, magnetoencephalography (MEG) has become a robust technique widely used in clinical medicine for the measurement and characterization of the magnetic fields induced by neuronal currents, for example, in an action potential. Powerful superconducting quantum interference devices (SQUIDs) are preferably employed for this purpose, thanks to the ultra-high sensitivity reachable by this type of magnetic sensors. MEG performs non-invasive and high-resolution brain mapping for disease diagnosis with a spatial resolution of a few millimeters and a temporal resolution of milliseconds, whereas an electroencephalogram (EEG) lacks spatial resolution but provides a satisfactory temporal resolution in the order of milliseconds [1]. MEG performed with SQUID devices exploits Josephson junctions to measure the magnetic flux at low temperature. Josephson junctions can record the minimum quantum variation of the magnetic flux, such as the magnetic quantum flux Φ [2]. SQUID sensors are particularly limited to operate by the low-temperature requirements leading to an overall bulky device with high maintenance costs. Other neuronal recording methods are also clinically used nowadays, such as functional magnetic resonance imaging (fMRI), EEG, and intracranial electroencephalography (iEEG). For instance, the fMRI technique is an indirect measurement of neuronal activities, which detects hemodynamic effects using blood oxygenation level-dependent responses [3,4]. The iEEG method measures the electrical activities in the brain through electrodes applied for functional cortex mapping [5]. However, at the current stage, the iEEG method is not compatible with magnetic resonance imaging (MRI).
In recent years, technological improvement has allowed researchers to come up with methods to dramatically increase the sensitivity of magnetic and optical sensors to the same level as SQUID magnetometers [6-8]. Nitrogen-vacancy (NV) center in diamond is a promising candidate for nanoscale sensing capable of detecting single neuronal activities. Nevertheless, voluminous optical devices are a major obstacle to transforming sensing platforms into miniaturized implantable and non-invasive wearable devices [9-14]. Alternatively, optical calcium imaging is another valuable technique to record brain cortical activity consisting of fluorescence imaging as an indicator of the presence of proteins in specific neuronal regions. Two-photon microscopy (TPEF) techniques are combined with calcium imaging to simultaneously record neuronal activities. The calcium concentration is related to electrical activities in neurons and optically detected through fluorescence-based calcium imaging with low temporal resolution in the order of millisecond range and spatial resolution of µm range [15-19].
On the other hand, wearable and implantable magnetic nanodevices can perform in vivo measurements at room temperature with a reduced cost and power consumption for biomedicalapplications ranging from cardiac monitoring to neuronal activity recording [3,20-25]. The recently emerging field of spintronic nanodevices exhibits sub-pT (sub-picotesla) detection capability for room-temperature applications and can be easily integrated on silicon substrates and flexible polymers [8,26–28]. Spintronic nanomagnetic devices are a promising candidate for implantable in vivo neuronal activity recording with high spatial and temporal resolution. Traditional microelectrodes relying on ionic processes for detecting action potentials can provoke an inflammatory response in adjacent biological tissues due to biofouling effects, resulting in reduced effectiveness and physical changes, such as ionic current contributions and a different impedance [29-31]. Unlike electrical implants, magnetic field sensors for neuronal activity recording do not require direct galvanic contact with the tissue since the magnetic field can penetrate inside the biological matter, thereby avoiding biofouling effects and allowing high endurance. It is foreseeable that magnetic field sensors will replace SQUID devices together with the current expensive cryogenic medical equipment for future portable heart and brain activity recording at room temperature.
We theoretically analyzed the different current sources in an activated neuron based on the Hodgkin-Huxley model (Section 2). Afterward, the theoretical intensity of currents flowing in an activated neuron is calculated using the NEURON software (Section 3.1). Based on the calculated current intensity, we then estimated the magnitude of the magnetic fields generated by a single axon according to the Biot-Savart law and classical electromagnetism (Section 3.2). In Sections 3.3 & 3.4, we theoretically calculated the magnetic field strength generated by the activity of individual neurons and neuronal networks at different distances. In addition, we commented on the feasibility of magnetic sensors for neuronal activity recording, especially referring to modern spintronic devices. The results reported in this work are corroborated and validated by Table 1. The purpose of this work is to provide peers with the expected theoretical value and experimental boundaries of the magnetic field produced by neuronal activities, as well as the feasibility of deploying different categories of magnetic field sensors for recording neuronal activities. Since the magnetic fields generated by neuronal activities are predominantly modulated by the protein channel dynamics and brain oscillations within 1 to 100 Hz, it is important to consider the sensitivity of magnetic sensors, particularly in the low-frequency regions 1 – 100 Hz, where the noise plays a more significant role [32].
| Table 1: Comparison of theoretical magnetic fields. | |||
| Neuron Model | Distance | Magnetic Fields | Reference |
| Single Neuron Electrical Stimulation | ~ 1 mm | ~ 2 fT | [52] |
| Giant Squid Axon | ~ 100 µm | ~ 600 pT | [9] |
| Neuronal Ensemble | ~ 100 µm | ~ 1 nT | [60] |
The NEURON software [33-36] is employed to calculate the total current induced in neurons during an action potential, which consists of the capacitive (Figure 1a), membrane (Figure 1c), and ionic currents (Figure 1e). Excluding secondary effects arising from frequency-dependent firing effects, the myelin sheath insulation, and axonal nonuniformities, such as the nodes of Ranvier [37-40], using the standard form of the Hodgkin-Huxley model [41], the following equivalence is valid:
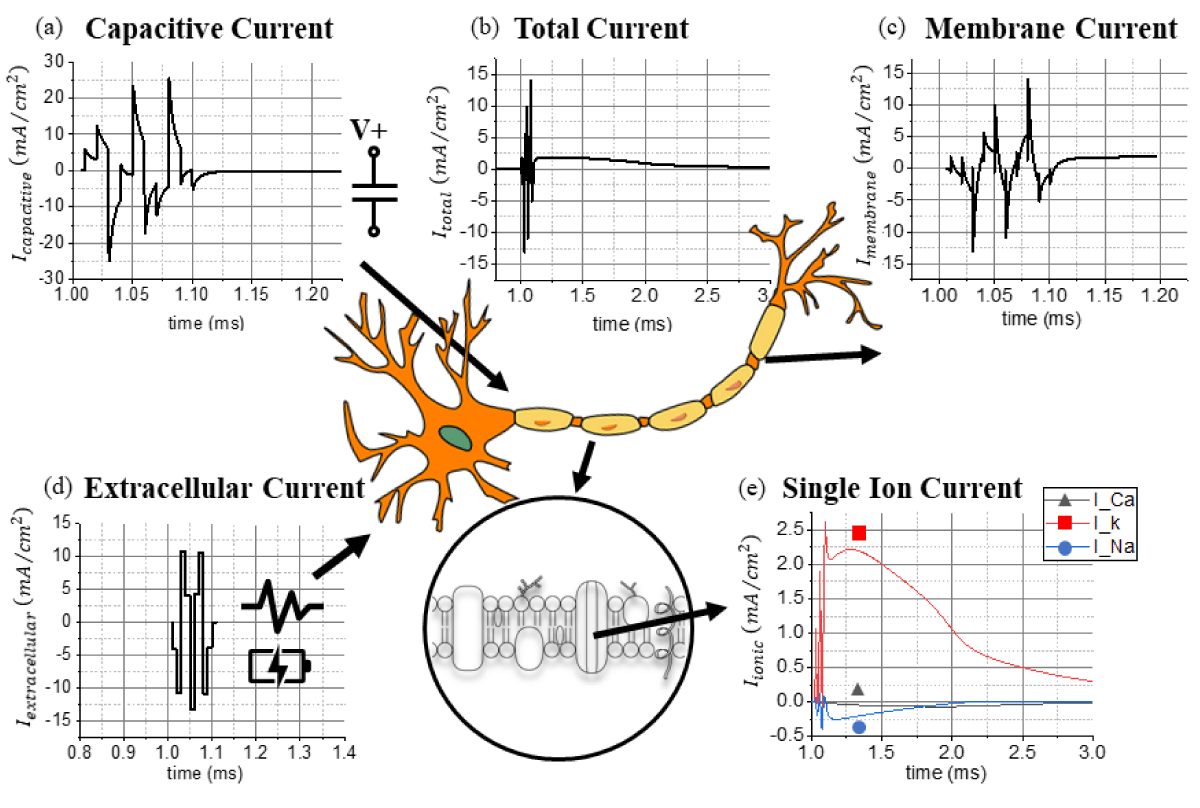
Figure 1: The extracted (a) capacitive current, (b) total current, (c) membrane current, (d) extracellular current, and (e) calcium, potassium, and sodium currents from a network of pyramidal neuronal cells using the NEURON software.
(1)
Where Cm is the membrane capacitance, Vm is the membrane voltage, Iex, is the extracellular current, and Iion is the ionic current. According to the Hodgkin-Huxley model for a giant squid axon, the ionic current is subdivided into three categories: sodium, potassium, and leakage current. The general expression for the ionic current contributions is stated by the following ordinary differential equations (ODEs) [41-44].
(2)
Where are the time and voltage-dependent rate constants correlated to each gate, and is a normalizing constant to express the maximum conductance once all the gates are open. Each channel (sodium Na+, potassium K+, and leakage l) is modeled with different numbers of gates, for example, 3-type-m and 1-type-h for the sodium channel. The probability of the channel opening is given by the generic equation [43]:
(3)
Where (1- pi) represents the probability that the i-gate is closed, and pi is the probability that the i-gate is open. The macroscopic total conductance Gi is dependent on all the open channels as [43]:
(4)
To derive the total current, the expression for the ionic current is substituted in (1):
(5)
The Hodgkin-Huxley model is strictly dependent on the conductance of each ionic channel. Therefore, an impedance variation within the biological tissue caused, for example, by electrical stimulation, generates a time-varying evolution of the system. The ionic current induced by transport phenomena across the ionic channels is responsible for generating electrochemical potentials. Furthermore, the membrane capacitance acts as a low-pass filter, and the dynamics of the biological system modulate the maximum cutoff frequency related to the frequency response of the ionic channels. For this reason, protein channels act as low-pass filters with respect to the dynamics of the current flowing in the system. Herein, dendrites, axons, and soma are affected by: the polarization effect [45-47], the orientation of the neuronal cells with respect to the induced current [48], and physical membrane parameters. The cell dynamics are affected by the local time-varying electric fields, which are used, for example, for cancer treatment [49]. The aforementioned ODE is solved in MATLAB and Simulink to characterize the behavior of single-neuron action potentials concerning external phenomena, such as microwave interactions with neuronal cells. Similar to electrical circuits, it is generally possible to correlate neuronal properties with their frequency response to express it using a circuit model. Since it is possible to measure the impedance characterizing neuronal activities, like in electrochemical systems, it is therefore possible to study the frequency response of neuronal activities. The working brain is also characterized by rhythmic activation and coupling of different resonance modes. Neurons work as notch filters excluding frequencies outside of the spectrum making the response to external current pulses dependent on several phenomena, such as the ionic (K+, Na+) membrane potential [50]. In addition to NEURON, other alternative common software includes: GENESIS, NEST, BRIAN, etc. As described by Tikidji-Hamburyan et al., each software uses peculiar properties for neuronal models, for example, including the conductance of ionic channels, single-compartment Hodgkin-Huxley model, extracellular chemical kinetics, diffusion model, and others [51].
Once the biophysical interpretation of different current sources in an activated neuron is expressed, different mathematical models are used to calculate and cross-validate the magnitude of the magnetic fields generated by the activated neurons [9,51,52]. Previous in vivo models are taken into consideration in order to evaluate the strength of magnetic fields induced by individual neurons [51,53]. The consistency of the FEM (finite element method) model is verified with magnetostatic simulations and corroborated with the LFPy open-source toolbox [51,52,54], which relies on the vector form of the Biot-Savart law. Other extrinsic phenomena can be taken into account, such as neuronal firing and calcium waves occurring when an ensemble of neuronal cells interacts together via action potentials [55].
Current induced in neurons during an action potential
Based on Saha et al.’s work on micromagnetic stimulation [48], the total induced current flowing in the soma during an action potential is calculated and validated with the NEURON software (Figure 1b & Table 2). MATLAB is applied for processing and visualizing different current sources, whereas COMSOL Multiphysics AC/DC Module is utilized to calculate the magnetic fields generated by a simplified axial structure (Figure 2a). Open-source computational neurobiology software relying on the NEURON software together with Python is used as a tool to reproduce the 3D magnetic field distribution around individual neurons during an evoked action potential [51,52]. Previous compatible models are taken into consideration, as well as a previously built customized geometry for the topological layout of neuronal cells in the hippocampal brain region. Figure 1e shows the theoretical magnitude of ionic currents arising from the calcium (Ca2+), potassium (K+), and sodium (Na+) potential. The ionic current flowing through calcium channels (Ca2+) is negligible compared to the potassium (K+) and sodium (Na+) channels. channels. Figure 1c represents the membrane current in the time domain extracted from the NEURON software as a solution for the theoretical ODE system. From 1.10 to 1.20 ms, the remaining current contribution is primarily attributed to the potassium (K+) channels, Figure 1d illustrates the extracellular current provoked by an external stimulus responsible for activating the synaptic response. The capacitive current (Figure 1a) is the contribution correlated to the time-dependent membrane potential in conjunction with the intrinsic capacitance of biological matter. This capacitive effect acts as a low-pass filtering effect around the neuronal cell and the axonal geometry. Summing together all the contributions extracted from the NEURON software, the theoretical model discussed above is applied to validate the simulation model proposed by Saha et al. [48] referring to neuronal networks. The source code of the standard geometry for the model is available in ModelDB [56] and published by Pashut et al. [57]..
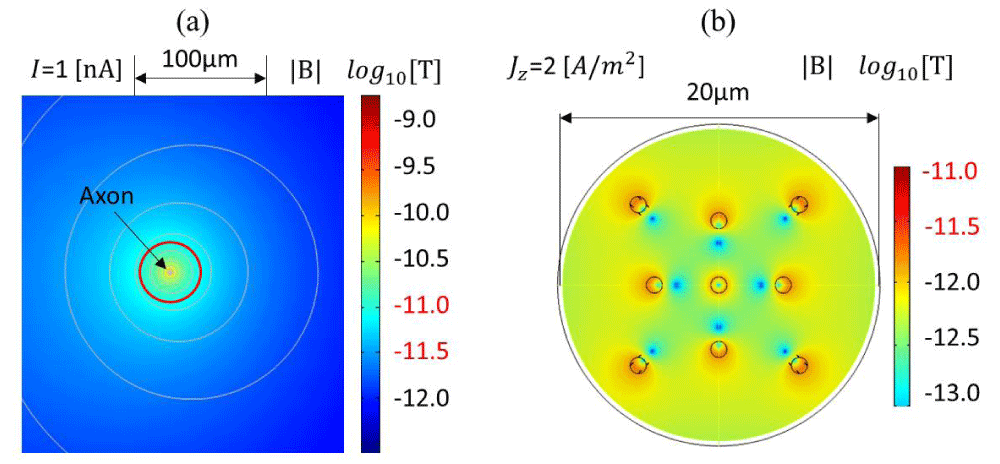
Figure 2: Contour plot of the calculated magnetic flux density in logarithmic scale generated with the COMSOL AC/DC Module based on a magnetostatic simulation: (a) assuming a 1 nA current flowing in a 0.1 µm diameter circular axon according to the parameters in Table 2, (b) assuming 2 A/m2 of current density, 1 µm circular axons, 1.081 kg/m3 density, and the physical parameters in Table 2, excluding the temperature variations.
| Table 2: Stimulation parameters for the NEURON software [33,48,56,57]. | |
| Current amplitude | 2-3 A |
| Duration | 0.4 ms |
| Resistance | 0.09 Ω |
| Inductance | 13 µH |
| Capacitance | 200 µF |
| Temperature | 34 °C |
Magnetic field generated by an axon
To emulate the magnetic field generated in neuronal cells, the axon is simplified as a single wire for a first-order approximation of the induced magnetic field according to the Biot-Savart law. The FEM simulation for a single axon is modeled as a single wire, as shown in Figure 2a. Using the Biot-Savart law, assuming I=1 nA, R=20 µm or 63 µm, and µ0=4π×10-7 H/m, the calculated magnetic flux densities are 10 pT and 3.16 pT, respectively. Figure 2a shows an axial cross-sectional view of the magnetic flux density around a simplified axonal circular geometry with a ϕ=0.1 µm diameter, when 1 nA of the total current is passing through it according to the parameters reported in Table 3. COMSOL Module AC/DC is used for the magnetostatic simulation. The resistivity of the biological tissue is extrapolated from white matter, ρ=5.8 Ω•m [58]. The relative permeability of the tissue is dominated by water (dielectric media, µr=1), whereas the conductivity σ is around 0.5 S/m. Figure 2b shows the magnetic field intensity for a circular axon of 1 µm diameter in the order of ~pT units, making it feasible for detecting single-neuron action potentials using micrometer spintronic nanomagnetic miniaturized sensors. Figure 2b illustrates the effect of a bundle of axons acting as a multipole, where the magnetic field cancels out in particular zones. A bundle of axons or neuronal cells can therefore amplify the total intensity of the magnetic field.
| Table 3: Parameters used in the simulation. | |
| Type of simulation | Magnetostatics |
| Relative permeability | 1 |
| Electrical conductivity | 0.5 S/m |
| Relative permittivity | 80 |
| Reference resistivity | 5.8 Ω·m |
| Resistivity temperature coefficient | 0 [1/K] |
| Reference temperature | 293 K |
| Microcoil radius | 0.1 µm |
Magnetic field generated by a single neuron
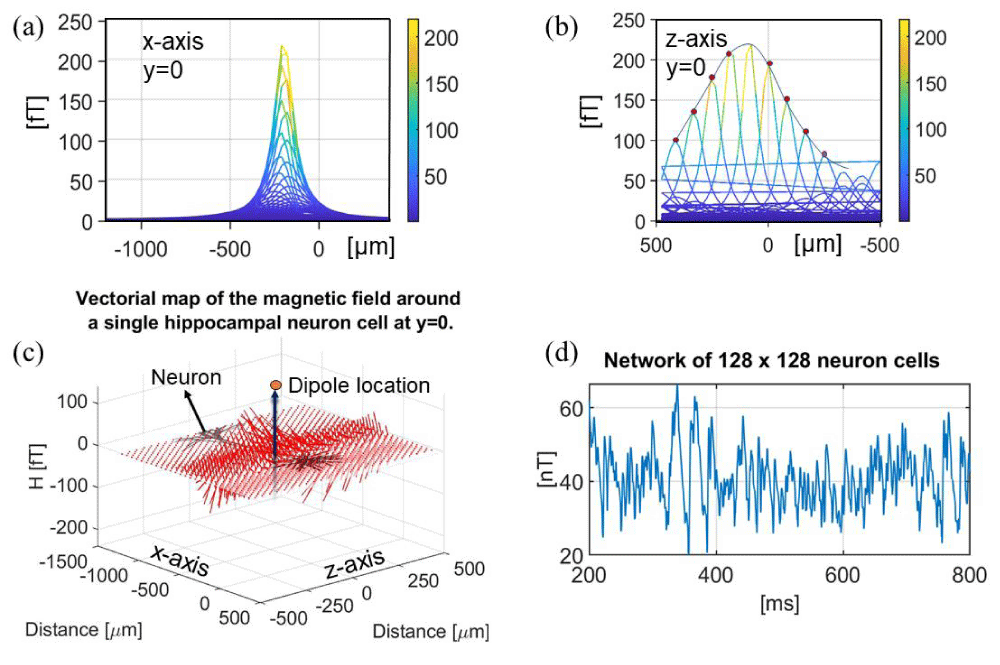
Using the LFPy software and open-source examples under the GNUv3 distribution (copyright 2017-2020 Computational Neuroscience Group NMBU) [51,52,54], the magnetic fields generated by individual neurons are calculated. The results show that the magnetic field at a ~1 µm distance from a dipole current source is in the order of 300 fT (femtotesla) as shown in Figure 3a,b. The parameters used in this simulation are listed in Table 4. The FEM simulations show the intensity of the magnetic field in the order of 1 pT for a similar synaptic current. This confirms that, at 1 µm distance, the magnetic field generated by an axon is in the order of ~pT units, which is also confirmed by Blagoev et al. [53]. Therefore, to record the magnetic fields generated by single neuronal activities, magnetic sensors should achieve sub-pT sensitivities due to the auto-cancellation induced by neighboring neurons together with low-pass filtering effects. Blagoev et al. [53] reported a similar estimation according to the LFPy results. The magnitude of the magnetic field generated by a single-neuron action potential is validated experimentally by Barry et al. on M. Infundibulum [9]. A theoretical calculation is presented in the supplementary materials (Figure S1). The in vivo maximum magnetic field measured with NV center in diamond sensors is in the order of 600 pT [9,59]. Therefore, the expectation for smaller neuronal cells being in the order of at least 1 pT is theoretically acceptable.
| Table 4: Parameters simulation Figure 3a-c from LFPy, ‘eegmegcalc.py’. | |
| Cell geometry | L5_Mainen96-LFPy.hoc |
| LFPy synapse weight value | 0.01 |
| LFPy synapse tau value | 5 |
| Dipole location | Mean on every axis |
| Type of simulation | MEG |
| Neuron cell position | {0, 0, 0} |
Figure 3: (a) Magnetic field intensity |H|, axial view (x-axis) for a neuronal cell for y = 0. (b) Magnetic field intensity |H|, axial view (z-axis) for y = 0. (c) Vectorial map of the magnetic field around a single hippocampal neuronal cell at y = 0. (d) Magnetic field intensity of 128×128 array of simplified neurons, single axon, pointing along the z-axis. Data was collected from LFPy under the GNU v3 license.
Magnetic field generated by neuronal networks
The magnetic fields generated by neuronal networks consisting of several hundreds of neurons have previously been calculated based on experimental data in the order of ~nT units, for example, in the hippocampal region [22,60-65]. Figure 3c schematically shows a simplified network consisting of 128 by 128 neurons aligned along the vertical axis (based on the LFPy example for a network of neuronal cells under the GNU license). The parameters used in this simulation are listed in Table 5. The simulated magnetic field concerning a simplified geometry is 40 – 50 nT at micrometer distance, which is one order of magnitude larger than the realistic values, 300 fT – 4.9 nT (9,52,53,60,64). The divergence is provoked by the absence of auto-cancellations among different compartments for this simplified model. In the LFPy software, the dipole is located at the centroid of the neuron geometry, which is averaged for each section in the NEURON software. Another effect to consider is that different contributions from individual segments cancel each other out, reducing the maximum detectable magnetic field. This is one limit of the computational NEURON simulation: in fact, the membrane acts as a low-pass filter that blocks the ionic transport phenomena. Another not well-quantifiable contribution is the dynamics of ionic channels, for example, the opening and closing states, which can affect the overall electrochemical potential and the neuronal ion transfer mediated by transport phenomena at the phase boundary interface, as described in the supplementary materials S2. Herein, the magnetic field generated by a single neuron is calculated to be in the order of 300 fT at ~1 – 5 µm distance.
| Table 5: Parameters simulation Figure 3d from LFPy ‘example_network.py’. | |
| Population parameters | |
| Radius | 200 µm |
| Population size | 128 ´ 128 Array |
| Network parameters | |
| Time resolution | 0.2 ms |
| Stop time | 800 ms |
| Equilibrium potential | - 65 mV |
| Temperature | 37 °C |
| Execution time | 4 min 35 s |
A few assumptions must be taken into consideration when evaluating the following results: the neuron is immersed in an infinite extracellular medium with homogeneous properties and independent of conductivity [54]; the magnetic field is generated by the dipole current via the Biot-Savart law that can be derived from the relativistic Maxwell-Heaviside equations considering negligible magnetic induction as H = (p×R)/4ΠR3, where p is the current dipole moment, R the vector between the dipole and the sensor location [54]. For a neuronal network, LFPy and NetPyNE can be successfully implemented if the cells have an elementary structure, and the single axon is elongated along the axis. In Figure 3a-c, the synaptic current is in the order of 0.2 nA – 0.4 nA according to the expectations for pyramidal neurons [66-68]. This confirms the right order of magnitude for the synaptic current evoked in a rodent pyramidal neuron. The theoretical calculations exhibit limitations arising from the curvature and axonal ramification, which results in the auto-cancellation of the magnetic field. External electromagnetic interference (EMI) can also lead to different intensities of the induced magnetic field. Parasitic eddy currents sum up to the total magnetic field contributions because of the skin effect provoked by the magnetization dynamics acting as mutual induction [69,71]. This effect has been considered to be negligible in the aforementioned simulation.
Magnetic sensor arrays for neuron activity recording
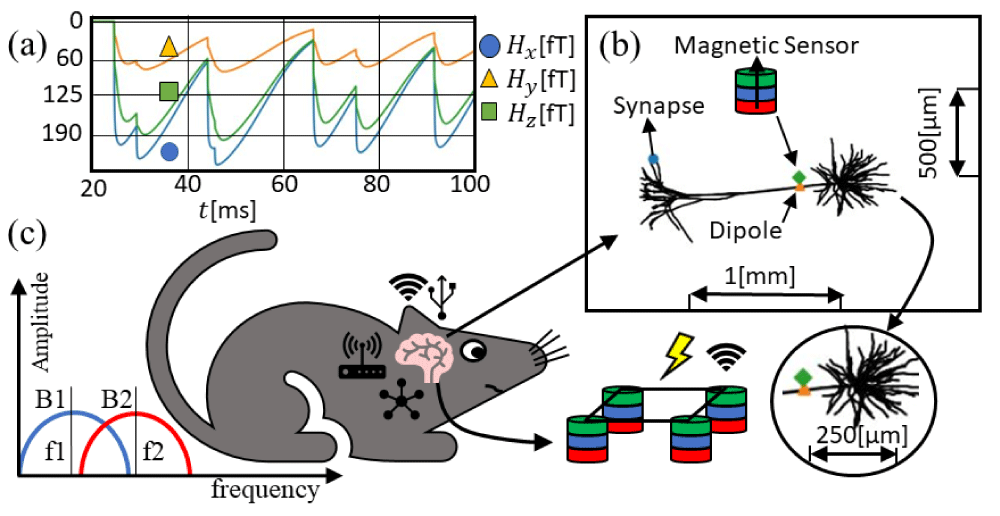
Detecting the magnetic fields generated by individual neurons with a microscale array of devices will allow the understanding of the basic building blocks for the development of neural networks leading to the full integration with machine-learning techniques for predicting wireless human-machine interfaces, as shown in Figure 4b. Stable and miniaturized spintronic nanodevices offer several advantages in comparison to other types of devices, such as room-temperature thermostability, high endurance, flexibility, non-invasive biocompatible coating to prevent corrosion and direct galvanic contact with brain tissues, robustness to external noise. Figure 4a represents the three components of the magnetic field for the three locations presented according to the LFPy example ‘eegmegcalc.py’ using the example geometry called ‘L5_Mainen96_LFPy.hoc’. The magnetic sensor is placed around 1 µm away from the dipole location. The location of the dipole is estimated as in Figure 3a-c averaging the position in the x-y plane for z=0. Figure 4c illustrates a frequency-selective filter used in combination with MTJ neural networks to extract particular frequencies depending on the biological system.
Figure 4: Magnetic field calculated in LFPy (a) applied magnetic field norm |H| in the rat brain, 2 µm from the dipole average per unit weight of every single segment in the NEURON software. (b) Representation of the pyramidal neuronal cell and the sensor location. (c) Representation of the mouse, where the network of single neurons is interconnected with nanoscale devices transmitting biological states. On the left, higher efficiency frequency-selective filters map out spatial frequencies concerning magnetic devices. On the bottom, a neural network of magnetic sensors is depicted.
Machine-learning techniques and toolboxes can be implemented to perform noise cancellations and improve the biophysical signals [72–77]. Machine-learning classification is a valid alternative to distinguish a real action potential to straightforwardly reduce its artifacts. Very often, artifacts can compromise the readability and the output signal in a way that is not predictable. AI and machine-learning techniques can play a key role in the optimization of neural networks and action potentials transferred along different pathways. Estimating the number of neurons can make it possible to correlate the brain power consumption for image processing computations and compare it with experimental low-power neural networks. Without machine-learning techniques and AI integration to instantly label positive action potentials, it will be complicated to create a human-machine interface toward gaining practical applications and an integrative understanding of neurobiological systems.
Having been developed and discovered in the 50s, single-neuronal activity recording with microelectrodes is not new [78]. Nowadays, technological improvement and information processing pave the way for the development of bidirectional closed-loop solutions for microelectrode sensing [79]. The spiking of neuronal activities in terms of action potentials is in the order of µV, easily detectable with present voltmeters and microelectrode arrays going down to nV resolution. The disadvantage is that electrode-based neuronal activity recording is affected by: corrosion phenomena at the interface, non-localized heating, and antenna effects, predominantly because the reaction at the interface is based on Nernstian electron transfer and not on the induced electromagnetic fields [80-89].
So far, the full form of the electromagnetic Maxwell-Heaviside equations has not been implemented for sensing single neuronal activities in terms of magnetic induction because of the low resolution of the induced magnetic field during action potentials. The control and leverage of the magnetic field inside the brain at low resolution would allow the regulation and sensing of spiking phenomena without involving on-chip electrochemical and electron transfer reactions, etc. [48].
The calculated magnetic fields are then compared and cross-validated with results published in the literature, which is summarized in Table 1. Hagen et al. estimated the magnetic field at 100 – 1000 µm scale in the order of ~fT units outside of the skull for electrical stimulation purposes [52]. For in vivo magnetic brain stimulation, it is possible to implant microscale devices in the order of 1 – 10 µm distance from the single neuron allowing for an enhanced magnetic field detection [48,90]. Furthermore, Hagen et al. estimated the magnetic field at the surface of the skull for networks of neuronal cells. It is reported that even at ~100 µm distance the magnetic field generated by neuronal networks is around ~100 fT [52,91]. In vivo measurements based on giant magnetoresistive (GMR) sensors can record magnetic fields generated from a neuronal ensemble in the order of ~nT units at a distance of ~100 µm and 100 pT at 1 µm [60]. Recent experimental results corroborate the theoretical results presented in Table 1, demonstrating the application of spintronic devices for single-neuron signal detection [92]. Therefore, the resolution of spintronic devices working on the principle of TMR (tunneling magnetoresistance) has reached a similar sensitivity to SQUID magnetometers for neuronal activity recording.
For neuroscientific applications, the analysis of single-neuron action potentials is important for the understanding of the interaction between neuronal signals. Spintronic magnetometers offer several advantages in comparison to SQUID devices, such as room-temperature thermostability without large bulky cryogenic instrumentations, and contactless spatial selectivity. A particular benefit of magnetic sensing is the removal of external ionic currents necessary for electrode-based methods, mitigating the risk of evoked perturbations. Since the magnetic field intensity for single neuronal activity is in the order of ~pT units or even ~fT units, ultra-high sensitivity magnetometers are required to extract neuronal activities from noisy environments. The magnetic field intensity induced by blood vessels and microfluidic dynamics is, at least, one hundred times more intense than a single-neuron activity. During in vivo experiments, the heart signal can be easily filtered in the frequency domain because of the characteristic spectrum.
Based on the results illustrated herein, the magnetic fields generated by single-neuron action potentials can be theoretically detected by ultra-high sensitivity magnetic field sensors. The calculations validate the feasibility of an array of implantable magnetic devices with sub-pT sensitivity to detect the magnetic field generated by a single-neuron action potential. Miniaturized nanoarrays of spintronic devices are ideal candidates to measure brain activities at a micrometer scale. Implantable nanosized spintronic sensors are a promising alternative for ultra-high sensitivity magnetic field detection in biocompatible nanodevices [99,100]. At present, atomic magnetometers are not enough miniaturized and robust to support in vivo implantation. Implantable nanoarrays of spintronic devices are compatible both in terms of achievable resolution at low frequencies [8], the optimal design for miniaturization, and the tremendous advantage of the magnetic-based device concerning external noise and stimuli, not to mention MRI scanning for a multitude of biocompatible coated magnetic sensors [93]. Currently, the sensitivity of spintronic sensors would enable the detection of a single-neuron magnetic field for magnetic brain mapping purposes [8,94–96]. Machine-learning techniques can be implemented to analyze neuronal signals and reject false positive action potentials, or validate specific action potentials via deep learning identification [97]. This would allow a statistical analysis of action potentials derived from biological neural networks. A similar approach is shown in Figure S3 of the supplementary materials.
This study was financially supported by the Minnesota Partnership for Biotechnology and Medical Genomics under Award Number ML2020. Chap 64. Art I, Section 4. The authors acknowledge the Robert F. Hartmann Chair Professorship, MN Drive Neuromodulation Fellowship, and Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this paper.
Notes
Copyright (C) 2012-2020 Computational Neuroscience Group, NMBU, under the terms of the GNU General Public License, version 3 or later. The code published in file ‘eegmegcalc.py’ available in the LFPy GitHub examples has been used for the magnetic field estimation, as well as the LFPy package under the GNU Public License GPLv3 distribution and added in the supplementary materials S4. The sensor locations have been changed accordingly and the neuron geometry has been set to the ‘L5_Mainen96_LFPy.hoc’. The code modifications are available in the supplementary material for the GNU Public License. Furthermore, the code published “example_network.py” has been used to extract magnetic field estimations for elementary neural networks.
Associated content
Supplementary materials are available: S1. Magnetic Fields Generated by Worm Axon; S2. Theoretical Electro-chemical Analysis for Brain sensing; S3. Graphical Interface; S4. Source Code LFPy simulation available on GitHub LFPy page under GNU license.
- Burle B, Spieser L, Roger C, Casini L, Hasbroucq T, Vidal F. Spatial and temporal resolutions of EEG: Is it really black and white? A scalp current density view. Int J Psychophysiol. 2015 Sep;97(3):210-20. doi: 10.1016/j.ijpsycho.2015.05.004. Epub 2015 May 12. PMID: 25979156; PMCID: PMC4548479.
- Clarke J. Squid Fundamentals. In: Weinstock H, editor. SQUID Sensors: Fundamentals, Fabrication and Applications. Dordrecht: Springer Netherlands; 1996 cited 2022 Apr;171–62. (NATO ASI Series). https://doi.org/10.1007/978-94-011-5674-5_1.
- Okada S, Bartelle BB, Li N, Breton-Provencher V, Lee JJ, Rodriguez E, Melican J, Sur M, Jasanoff A. Calcium-dependent molecular fMRI using a magnetic nanosensor. Nat Nanotechnol. 2018 Jun;13(6):473-477. doi: 10.1038/s41565-018-0092-4. Epub 2018 Apr 30. PMID: 29713073; PMCID: PMC6086382.
- Caparelli EC, Zhai T, Yang Y. Simultaneous Transcranial Magnetic Stimulation and Functional Magnetic Resonance Imaging: Aspects of Technical Implementation. Front Neurosci. 2020 cited 2022 Apr 17; 14. https://www.frontiersin.org/article/10.3389/fnins.2020.554714
- Wang Y, Yan J, Wen J, Yu T, Li X. An Intracranial Electroencephalography (iEEG) Brain Function Mapping Tool with an Application to Epilepsy Surgery Evaluation. Front Neuroinform. 2016 Apr 25;10:15. doi: 10.3389/fninf.2016.00015. PMID: 27199729; PMCID: PMC4842770.
- PicoTesla magnetic tunneling junction sensors integrated with double staged magnetic flux concentrators: Applied Physics Letters: 2022 Jan 8; 113: 24. https://aip.scitation.org/doi/10.1063/1.5052355
- Chaves RC, Freitas PP. Low frequency picotesla field detection using hybrid MgO based tunnel sensors: Applied Physics Letters. 2022 Jan 8; 91:10. https://aip.scitation.org/doi/10.1063/1.2775802
- Oogane M, Fujiwara K, Kanno A, Nakano T, Wagatsuma H, Arimoto T. Sub-pT magnetic field detection by tunnel magneto-resistive sensors. Appl Phys Express. 2021 Nov; 14(12):123002.
- Barry JF, Turner MJ, Schloss JM, Glenn DR, Song Y, Lukin MD, Park H, Walsworth RL. Optical magnetic detection of single-neuron action potentials using quantum defects in diamond. Proc Natl Acad Sci U S A. 2016 Dec 6;113(49):14133-14138. doi: 10.1073/pnas.1601513113. Epub 2016 Nov 22. Erratum in: Proc Natl Acad Sci U S A. 2017 Aug 7;: PMID: 27911765; PMCID: PMC5150388.
- Schirhagl R, Chang K, Loretz M, Degen CL. Nitrogen-vacancy centers in diamond: nanoscale sensors for physics and biology. Annu Rev Phys Chem. 2014;65:83-105. doi: 10.1146/annurev-physchem-040513-103659. Epub 2013 Nov 21. PMID: 24274702.
- Savukov I, Kim YJ, Schultz G. Detection of ultra-low field NMR signal with a commercial QuSpin single-beam atomic magnetometer. J Magn Reson. 2020 Aug;317:106780. doi: 10.1016/j.jmr.2020.106780. Epub 2020 Jul 12. PMID: 32688163.
- Boto E, Meyer SS, Shah V, Alem O, Knappe S, Kruger P, Fromhold TM, Lim M, Glover PM, Morris PG, Bowtell R, Barnes GR, Brookes MJ. A new generation of magnetoencephalography: Room temperature measurements using optically-pumped magnetometers. Neuroimage. 2017 Apr 1;149:404-414. doi: 10.1016/j.neuroimage.2017.01.034. Epub 2017 Jan 25. PMID: 28131890; PMCID: PMC5562927.
- Hill RM, Boto E, Holmes N, Hartley C, Seedat ZA, Leggett J, Roberts G, Shah V, Tierney TM, Woolrich MW, Stagg CJ, Barnes GR, Bowtell R, Slater R, Brookes MJ. A tool for functional brain imaging with lifespan compliance. Nat Commun. 2019 Nov 5;10(1):4785. doi: 10.1038/s41467-019-12486-x. Erratum in: Nat Commun. 2019 Dec 4;10(1):5628. PMID: 31690797; PMCID: PMC6831615.
- Webb JL, Troise L, Hansen NW, Achard J, Brinza O, Staacke R. Optimization of a Diamond Nitrogen Vacancy Centre Magnetometer for Sensing of Biological Signals. Front Phys. 2020 cited 2022 Jun 19; 8. https://www.frontiersin.org/article/10.3389/fphy.2020.522536
- Song A, Gauthier JL, Pillow JW, Tank DW, Charles AS. Neural anatomy and optical microscopy (NAOMi) simulation for evaluating calcium imaging methods. J Neurosci Methods. 2021 Jul 1; 358: 109173. doi: 10.1016/j.jneumeth.2021.109173. Epub 2021 Apr 8. PMID: 33839190; PMCID: PMC8217135.
- Reynolds JP, Zheng K, Rusakov DA. Multiplexed calcium imaging of single-synapse activity and astroglial responses in the intact brain. Neurosci Lett. 2019 Jan 10; 689: 26-32. doi: 10.1016/j.neulet.2018.06.024. Epub 2018 Jun 19. PMID: 29908948; PMCID: PMC6335263.
- Linden NJ, Tabuena DR, Steinmetz NA, Moody WJ, Brunton SL, Brunton BW. Go with the FLOW: visualizing spatiotemporal dynamics in optical widefield calcium imaging. J R Soc Interface. 2021 Aug;18(181):20210523. doi: 10.1098/rsif.2021.0523. Epub 2021 Aug 25. PMID: 34428947; PMCID: PMC8385384.
- Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007 Oct 4;56(1):43-57. doi: 10.1016/j.neuron.2007.08.003. PMID: 17920014; PMCID: PMC2268027.
- .Rochefort NL, Jia H, Konnerth A. Calcium imaging in the living brain: prospects for molecular medicine. Trends Mol Med. 2008 Sep;14(9):389-99. doi: 10.1016/j.molmed.2008.07.005. Epub 2008 Aug 12. PMID: 18701348.
- Gao L, Wang J, Guan S, Du M, Wu K, Xu K, Zou L, Tian H, Fang Y. Magnetic Actuation of Flexible Microelectrode Arrays for Neural Activity Recordings. Nano Lett. 2019 Nov 13;19(11):8032-8039. doi: 10.1021/acs.nanolett.9b03232. Epub 2019 Oct 8. PMID: 31580687.
- Barbieri F, Trauchessec V, Caruso L, Trejo-Rosillo J, Telenczuk B, Paul E, Bal T, Destexhe A, Fermon C, Pannetier-Lecoeur M, Ouanounou G. Local recording of biological magnetic fields using Giant Magneto Resistance-based micro-probes. Sci Rep. 2016 Dec 19; 6:39330. doi: 10.1038/srep39330. PMID: 27991562; PMCID: PMC5171880.
- Pannetier-Lecoeur M, Polovy H, Sergeeva-Chollet N, Cannies G, Fermon C, Parkkonen L. Magnetocardiography with GMR-based sensors. J Phys Conf Ser. 2011 Jul; 303: 012054.
- Pannetier-Lecoeur M, Fermon C, Polovy H, Dyvorne H, Sergeeva-Chollet N, Paul J. GMR-based sensors for ultra-sensitive magnetometry. In: 2009 IEEE SENSORS. 2009; 1856–9.
- Pannetier M, Fermon C, Legoff G, Simola J, Kerr E, Welling M. Ultra-sensitive field sensors - an alternative to SQUIDs. IEEE Trans Appl Supercond. 2005 Jun; 15(2):892–5.
- Zuo S, Heidari H, Farina D, Nazarpour K. Miniaturized Magnetic Sensors for Implantable Magnetomyography. Adv Mater Technol. 2020; 5(6):2000185.
- Chen X, Mi W. Mechanically tunable magnetic and electronic transport properties of flexible magnetic films and their heterostructures for spintronics. J Mater Chem C. 2021; 9(30):9400–30.
- Amara S, Sevilla GAT, Hawsawi M, Mashraei Y, Mohammed H, Cruz ME. High-Performance Flexible Magnetic Tunnel Junctions for Smart Miniaturized Instruments. Adv Eng Mater. 2018; 20(10):1800471.
- Cardoso S, Leitao DC, Dias TM, Valadeiro J, Silva MD, Chicharo A. Challenges and trends in magnetic sensor integration with microfluidics for biomedical applications. J Phys Appl Phys. 2017 Apr; 50(21):213001.
- Hanssen BL, Siraj S, Wong DKY. Recent strategies to minimise fouling in electrochemical detection systems. Rev Anal Chem. 2016 Apr 1; 35(1):1–28.
- Seaton BT, Hill DF, Cowen SL, Heien ML. Mitigating the Effects of Electrode Biofouling-Induced Impedance for Improved Long-Term Electrochemical Measurements in Vivo. Anal Chem. 2020 May 5;92(9):6334-6340. doi: 10.1021/acs.analchem.9b05194. Epub 2020 Apr 16. PMID: 32298105; PMCID: PMC8281608.
- Kousar A, Peltola E, Laurila T. Nanostructured Geometries Strongly Affect Fouling of Carbon Electrodes. ACS Omega. 2021 Sep 29;6(40):26391-26403. doi: 10.1021/acsomega.1c03666. PMID: 34660997; PMCID: PMC8515610.
- Mably AJ, Colgin LL. Gamma oscillations in cognitive disorders. Curr Opin Neurobiol. 2018 Oct;52:182-187. doi: 10.1016/j.conb.2018.07.009. Epub 2018 Aug 16. PMID: 30121451; PMCID: PMC6139067.
- Carnevale T, Hines M, Hines M, Hines M. The NEURON Book. Cambridge University Press. 2006.
- Carnevale, Hines ML, Hines ML, Hines ML. The NEURON simulation environment in epilepsy research. London: Elsevier. 2009.
- Hines ML, Carnevale NT. The NEURON simulation environment. Neural Comput. 1997 Aug 15; 9(6):1179-209. doi: 10.1162/neco.1997.9.6.1179. PMID: 9248061.
- Hines M. The neurosimulator NEURON. Methods Neuronal Model Ed C Koch Segev Camb MA MIT Press. 1998; 129-136.
- Miller RN, Rinzel J. The dependence of impulse propagation speed on firing frequency, dispersion, for the Hodgkin-Huxley model. Biophys J. 1981 May;34(2):227-59. doi: 10.1016/S0006-3495(81)84847-3. PMID: 7236850; PMCID: PMC1327469.
- Lee SG, Neiman A, Kim S. Coherence resonance in a Hodgkin-Huxley neuron. Phys Rev E. 1998 Mar 1; 57(3):3292–7.
- Ford MC, Alexandrova O, Cossell L, Stange-Marten A, Sinclair J, Kopp-Scheinpflug C, Pecka M, Attwell D, Grothe B. Tuning of Ranvier node and internode properties in myelinated axons to adjust action potential timing. Nat Commun. 2015 Aug 25;6:8073. doi: 10.1038/ncomms9073. PMID: 26305015; PMCID: PMC4560803.
- Susuki K. Node of Ranvier disruption as a cause of neurological diseases. ASN Neuro. 2013 Aug 7;5(3):209-19. doi: 10.1042/AN20130025. PMID: 23834220; PMCID: PMC3736360.
- HODGKIN AL, HUXLEY AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952 Aug;117(4):500-44. doi: 10.1113/jphysiol.1952.sp004764. PMID: 12991237; PMCID: PMC1392413.
- Clay JR, Paydarfar D, Forger DB. A simple modification of the Hodgkin and Huxley equations explains type 3 excitability in squid giant axons. J R Soc Interface. 2008 Dec 6;5(29):1421-8. doi: 10.1098/rsif.2008.0166. PMID: 18544505; PMCID: PMC2607356.
- Siciliano R. The Hodgkin-Huxley Model. 41.
- Colwell LJ, Brenner MP. Action potential initiation in the hodgkin-huxley model. PLoS Comput Biol. 2009 Jan;5(1):e1000265. doi: 10.1371/journal.pcbi.1000265. Epub 2009 Jan 16. PMID: 19148265; PMCID: PMC2607014.
- Albertson AJ, Williams SB, Hablitz JJ. Regulation of epileptiform discharges in rat neocortex by HCN channels. J Neurophysiol. 2013 Oct;110(8):1733-43. doi: 10.1152/jn.00955.2012. Epub 2013 Jul 17. PMID: 23864381; PMCID: PMC3798942.
- Bikson M, Inoue M, Akiyama H, Deans JK, Fox JE, Miyakawa H, Jefferys JG. Effects of uniform extracellular DC electric fields on excitability in rat hippocampal slices in vitro. J Physiol. 2004 May 15;557(Pt 1):175-90. doi: 10.1113/jphysiol.2003.055772. Epub 2004 Feb 20. PMID: 14978199; PMCID: PMC1665051.
- Chan CY, Hounsgaard J, Nicholson C. Effects of electric fields on transmembrane potential and excitability of turtle cerebellar Purkinje cells in vitro. J Physiol. 1988 Aug;402:751-71. doi: 10.1113/jphysiol.1988.sp017232. PMID: 3236254; PMCID: PMC1191919.
- Saha R, Faramarzi S, Bloom RP, Benally OJ, Wu K, di Girolamo A. Strength-frequency curve for micromagnetic neurostimulation through EPSPs on rat hippocampal neurons and numerical modeling of magnetic microcoil. 2021 Dec [cited 2022 Jan 7] 2021.11.30.470598. https://www.biorxiv.org/content/10.1101/2021.11.30.470598v1
- Delso C, Martínez JM, Cebrián G, Álvarez I, Raso J. Understanding the occurrence of tailing in survival curves of Salmonella Typhimurium treated by pulsed electric fields. Bioelectrochemistry. 2020 Oct;135:107580. doi: 10.1016/j.bioelechem.2020.107580. Epub 2020 Jun 2. PMID: 32526677.
- Hutcheon B, Yarom Y. Resonance, oscillation and the intrinsic frequency preferences of neurons. Trends Neurosci. 2000 May;23(5):216-22. doi: 10.1016/s0166-2236(00)01547-2. PMID: 10782127.
- Tikidji-Hamburyan RA, Narayana V, Bozkus Z, El-Ghazawi TA. Software for Brain Network Simulations: A Comparative Study. Front Neuroinformatics. 2017 [cited 2022 Dec 29]; 11. https://www.frontiersin.org/articles/10.3389/fninf.2017.00046
- Hagen E, Næss S, Ness TV, Einevoll GT. Multimodal Modeling of Neural Network Activity: Computing LFP, ECoG, EEG, and MEG Signals With LFPy 2.0. Front Neuroinform. 2018 Dec 18;12:92. doi: 10.3389/fninf.2018.00092. PMID: 30618697; PMCID: PMC6305460.
- Lindén H, Hagen E, Lęski S, Norheim ES, Pettersen KH, Einevoll GT. LFPy: a tool for biophysical simulation of extracellular potentials generated by detailed model neurons. Front Neuroinform. 2014 Jan 16;7:41. doi: 10.3389/fninf.2013.00041. PMID: 24474916; PMCID: PMC3893572.
- Blagoev KB, Mihaila B, Travis BJ, Alexandrov LB, Bishop AR, Ranken D, Posse S, Gasparovic C, Mayer A, Aine CJ, Ulbert I, Morita M, Müller W, Connor J, Halgren E. Modelling the magnetic signature of neuronal tissue. Neuroimage. 2007 Aug 1;37(1):137-48. doi: 10.1016/j.neuroimage.2007.04.033. Epub 2007 May 4. PMID: 17544300.
- Hagen E, Ness TV. LFPy. 2021. https://github.com/LFPy/LFPy
- Hounsgaard J, Kiehn O. Calcium spikes and calcium plateaux evoked by differential polarization in dendrites of turtle motoneurones in vitro. J Physiol. 1993 Aug;468:245-59. doi: 10.1113/jphysiol.1993.sp019769. PMID: 8254508; PMCID: PMC1143824.
- McDougal RA, Morse TM, Carnevale T, Marenco L, Wang R, Migliore M, Miller PL, Shepherd GM, Hines ML. Twenty years of ModelDB and beyond: building essential modeling tools for the future of neuroscience. J Comput Neurosci. 2017 Feb;42(1):1-10. doi: 10.1007/s10827-016-0623-7. Epub 2016 Sep 15. PMID: 27629590; PMCID: PMC5279891.
- Pashut T, Wolfus S, Friedman A, Lavidor M, Bar-Gad I, Yeshurun Y, Korngreen A. Mechanisms of magnetic stimulation of central nervous system neurons. PLoS Comput Biol. 2011 Mar;7(3):e1002022. doi: 10.1371/journal.pcbi.1002022. Epub 2011 Mar 24. PMID: 21455288; PMCID: PMC3063755.
- Li CL, Bak AF, Parker LO. Specific resistivity of the cerebral cortex and white matter. Exp Neurol. 1968 Apr;20(4):544-57. doi: 10.1016/0014-4886(68)90108-8. PMID: 5659447.
- Glenn DR, Lee K, Park H, Weissleder R, Yacoby A, Lukin MD, Lee H, Walsworth RL, Connolly CB. Single-cell magnetic imaging using a quantum diamond microscope. Nat Methods. 2015 Aug;12(8):736-738. doi: 10.1038/nmeth.3449. Epub 2015 Jun 22. PMID: 26098019; PMCID: PMC4521973.
- Caruso L, Wunderle T, Lewis CM, Valadeiro J, Trauchessec V, Trejo Rosillo J, Amaral JP, Ni J, Jendritza P, Fermon C, Cardoso S, Freitas PP, Fries P, Pannetier-Lecoeur M. In Vivo Magnetic Recording of Neuronal Activity. Neuron. 2017 Sep 13;95(6):1283-1291.e4. doi: 10.1016/j.neuron.2017.08.012. Epub 2017 Aug 30. PMID: 28844526; PMCID: PMC5744593.
- Amaral J, Cardoso S, Freitas PP, Sebastião AM. Toward a system to measure action potential on mice brain slices with local magnetoresistive probes. J Appl Phys. 2011 Apr 1;109(7):07B308.
- Fujiwara K, Oogane M, Kanno A, Imada M, Jono J, Terauchi T. Magnetocardiography and magnetoencephalography measurements at room temperature using tunnel magneto-resistance sensors. Appl Phys Express. 2018 Jan 18; 11(2):023001.
- Pannetier-Lecoeur M, Parkkonen L, Sergeeva-Chollet N, Polovy H, Fermon C, Fowley C. Magnetocardiography with sensors based on giant magnetoresistance. Appl Phys Lett. 2011 Apr 11; 98(15):153705.
- Chopin C, Torrejon J, Solignac A, Fermon C, Jendritza P, Fries P, Pannetier-Lecoeur M. Magnetoresistive Sensor in Two-Dimension on a 25 μm Thick Silicon Substrate for In Vivo Neuronal Measurements. ACS Sens. 2020 Nov 25;5(11):3493-3500. doi: 10.1021/acssensors.0c01578. Epub 2020 Oct 27. PMID: 33108725.
- Guedes A, Macedo R, Jaramillo G, Cardoso S, Freitas PP, Horsley DA. Hybrid GMR Sensor Detecting 950 pT/sqrt(Hz) at 1 Hz and Room Temperature. Sensors (Basel). 2018 Mar 6;18(3):790. doi: 10.3390/s18030790. PMID: 29509677; PMCID: PMC5876515.
- Dura-Bernal S, Suter BA, Gleeson P, Cantarelli M, Quintana A, Rodriguez F, Kedziora DJ, Chadderdon GL, Kerr CC, Neymotin SA, McDougal RA, Hines M, Shepherd GM, Lytton WW. NetPyNE, a tool for data-driven multiscale modeling of brain circuits. Elife. 2019 Apr 26;8:e44494. doi: 10.7554/eLife.44494. PMID: 31025934; PMCID: PMC6534378.
- Lytton WW, Seidenstein AH, Dura-Bernal S, McDougal RA, Schürmann F, Hines ML. Simulation Neurotechnologies for Advancing Brain Research: Parallelizing Large Networks in NEURON. Neural Comput. 2016 Oct;28(10):2063-90. doi: 10.1162/NECO_a_00876. Epub 2016 Aug 24. PMID: 27557104; PMCID: PMC5295685.
- Hammond C, Goaillard JM, Debanne D, Gaiarsa JL. Chapter 18 - Synaptic plasticity. In: Hammond C, editor. Cellular and Molecular Neurophysiology Fourth Edition. Boston: Academic Press. 2015 [cited 2022 Feb 5]; 361–89. https://www.sciencedirect.com/science/article/pii/B9780123970329000182
- Lu HY, Zhu JG, Ramsden VS, Hui SYR. Measurement and modeling of stray capacitances in high frequency transformers. In: 30th Annual IEEE Power Electronics Specialists Conference Record (Cat No99CH36321). 1999; 763–8 .2.
- Cheng Y, Wang G, Ghovanloo M. Analytical Modeling and Optimization of Small Solenoid Coils for Millimeter-Sized Biomedical Implants. IEEE Trans Microw Theory Tech. 2017 Mar;65(3):1024–35.
- Wu J, Quinn V, Bernstein GH. An Inductive Link with Integrated Receiving Coil-Coupling Coefficient and Link Efficiency. J Comput Electron. 2005 Dec 1;4(3):221–30.
- Houston B, Thompson M, Ko A, Chizeck H. A machine-learning approach to volitional control of a closed-loop deep brain stimulation system. J Neural Eng. 2019 Feb;16(1):016004. doi: 10.1088/1741-2552/aae67f. Epub 2018 Nov 16. PMID: 30444218.
- Neumann WJ, Rodriguez-Oroz MC. Machine Learning Will Extend the Clinical Utility of Adaptive Deep Brain Stimulation. Mov Disord. 2021 Apr; 36(4):796-799. doi: 10.1002/mds.28567. PMID: 33851753.
- Boutet A, Madhavan R, Elias GJB, Joel SE, Gramer R, Ranjan M, Paramanandam V, Xu D, Germann J, Loh A, Kalia SK, Hodaie M, Li B, Prasad S, Coblentz A, Munhoz RP, Ashe J, Kucharczyk W, Fasano A, Lozano AM. Predicting optimal deep brain stimulation parameters for Parkinson's disease using functional MRI and machine learning. Nat Commun. 2021 May 24;12(1):3043. doi: 10.1038/s41467-021-23311-9. PMID: 34031407; PMCID: PMC8144408.
- Shamir RR, Duchin Y, Kim J, Patriat R, Marmor O, Bergman H, Vitek JL, Sapiro G, Bick A, Eliahou R, Eitan R, Israel Z, Harel N. Microelectrode Recordings Validate the Clinical Visualization of Subthalamic-Nucleus Based on 7T Magnetic Resonance Imaging and Machine Learning for Deep Brain Stimulation Surgery. Neurosurgery. 2019 Mar 1;84(3):749-757. doi: 10.1093/neuros/nyy212. PMID: 29800386; PMCID: PMC6500885.
- Bermudez C, Rodriguez W, Huo Y, Hainline AE, Li R, Shults R. Towards machine learning prediction of deep brain stimulation (DBS) intra-operative efficacy maps. In: Medical Imaging 2019: Image Processing. SPIE; 2019 [cited 2022 Jan 8]; 528–34. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10949/1094922/Towards-machine-learning-prediction-of-deep-brain-stimulation-DBS-intra/10.1117/12.2509728.full
- Machine Learning Approach to Optimizing Combined Stimulation and Medication Therapies for Parkinson’s Disease - ScienceDirect. Cited 2022 Jan 8. https://www.sciencedirect.com/science/article/pii/S1935861X15010049
- Dowben RM, Rose JE. A metal-filled microelectrode. Science. 1953 Jul 3; 118(3053):22-4. doi: 10.1126/science.118.3053.22. PMID: 13076162.
- Topalovic U, Barclay S, Ling C, Alzuhair A, Yu W, Hokhikyan V. A wearable platform for closed-loop stimulation and recording of single- neuron and local field potential activity in freely-moving humans. bioRxiv; 2022 cited 2022 Feb 9;2022.02.05.479253. https://www.biorxiv.org/content/10.1101/2022.02.05.479253v1
- Cogan SF. Neural stimulation and recording electrodes. Annu Rev Biomed Eng. 2008; 10:275-309. doi: 10.1146/annurev.bioeng.10.061807.160518. PMID: 18429704.
- Cogan SF, Ehrlich J, Plante TD, Smirnov A, Shire DB, Gingerich M, Rizzo JF. Sputtered iridium oxide films for neural stimulation electrodes. J Biomed Mater Res B Appl Biomater. 2009 May;89(2):353-361. doi: 10.1002/jbm.b.31223. PMID: 18837458; PMCID: PMC7442142.
- Park HJ, Bonmassar G, Kaltenbach JA, Machado AG, Manzoor NF, Gale JT. Activation of the central nervous system induced by micro-magnetic stimulation. Nat Commun. 2013;4:2463. doi: 10.1038/ncomms3463. PMID: 24030203; PMCID: PMC3845906.
- Mohsin SA, Sheikh NM, Saeed U. MRI-induced heating of deep brain stimulation leads. Phys Med Biol. 2008 Oct 21;53(20):5745-56. doi: 10.1088/0031-9155/53/20/012. Epub 2008 Sep 26. PMID: 18824784.
- Gregersen F, Göksu C, Schaefers G, Xue R, Thielscher A, Hanson LG. Safety evaluation of a new setup for transcranial electric stimulation during magnetic resonance imaging. Brain Stimul. 2021 May-Jun;14(3):488-497. doi: 10.1016/j.brs.2021.02.019. Epub 2021 Mar 9. PMID: 33706007.
- Angelone LM, Ahveninen J, Belliveau JW, Bonmassar G. Analysis of the role of lead resistivity in specific absorption rate for deep brain stimulator leads at 3T MRI. IEEE Trans Med Imaging. 2010 Apr;29(4):1029-38. doi: 10.1109/TMI.2010.2040624. Epub 2010 Mar 22. PMID: 20335090; PMCID: PMC3145199.
- Golestanirad L, Kazemivalipour E, Lampman D, Habara H, Atalar E, Rosenow J, Pilitsis J, Kirsch J. RF heating of deep brain stimulation implants in open-bore vertical MRI systems: A simulation study with realistic device configurations. Magn Reson Med. 2020 Jun;83(6):2284-2292. doi: 10.1002/mrm.28049. Epub 2019 Nov 2. PMID: 31677308; PMCID: PMC7047541.
- Rahimpour S, Kiyani M, Hodges SE, Turner DA. Deep brain stimulation and electromagnetic interference. Clin Neurol Neurosurg. 2021 Apr;203:106577. doi: 10.1016/j.clineuro.2021.106577. Epub 2021 Feb 25. PMID: 33662743; PMCID: PMC8081063.
- Oathes DJ, Balderston NL, Kording KP, DeLuisi JA, Perez GM, Medaglia JD, Fan Y, Duprat RJ, Satterthwaite TD, Sheline YI, Linn KA. Combining transcranial magnetic stimulation with functional magnetic resonance imaging for probing and modulating neural circuits relevant to affective disorders. Wiley Interdiscip Rev Cogn Sci. 2021 Jul;12(4):e1553. doi: 10.1002/wcs.1553. Epub 2021 Jan 19. PMID: 33470055; PMCID: PMC8521438.
- Doering M, Kieninger J, Urban GA, Weltin A. Electrochemical microelectrode degradation monitoring:in situinvestigation of platinum corrosion at neutral pH. J Neural Eng. 2022 Jan 24;19(1). doi: 10.1088/1741-2552/ac47da. PMID: 34983028.
- Kim C, Nichols E, Kim BN. Choosing the Optimal Power Coils Using Open-Source k-Oriented Design Automation. IEEE Trans Biomed Circuits Syst. 2021 Feb;15(1):159-170. doi: 10.1109/TBCAS.2021.3059934. Epub 2021 Mar 30. PMID: 33729948.
- Salmelin R, Hari R, Lounasmaa OV, Sams M. Dynamics of brain activation during picture naming. Nature. 1994 Mar 31;368(6470):463-5. doi: 10.1038/368463a0. PMID: 8133893.
- Kanno A, Nakasato N, Oogane M, Fujiwara K, Nakano T, Arimoto T, Matsuzaki H, Ando Y. Scalp attached tangential magnetoencephalography using tunnel magneto-resistive sensors. Sci Rep. 2022 Apr 12;12(1):6106. doi: 10.1038/s41598-022-10155-6. PMID: 35414691; PMCID: PMC9005603.
- Zhang Y, Le S, Li H, Ji B, Wang MH, Tao J, Liang JQ, Zhang XY, Kang XY. MRI magnetic compatible electrical neural interface: From materials to application. Biosens Bioelectron. 2021 Dec 15;194:113592. doi: 10.1016/j.bios.2021.113592. Epub 2021 Sep 1. PMID: 34507098.
- Shirotori S, Kikitsu A, Higashi Y, Kurosaki Y, Iwasaki H. Symmetric Response Magnetoresistance Sensor With Low 1/f Noise by Using an Antiphase AC Modulation Bridge. IEEE Trans Magn. 2021 Feb;57(2):1–5.
- Bonmassar G, Lee SW, Freeman DK, Polasek M, Fried SI, Gale JT. Microscopic magnetic stimulation of neural tissue. Nat Commun. 2012 Jun 26;3:921. doi: 10.1038/ncomms1914. PMID: 22735449; PMCID: PMC3621430.
- Goyal A, Goetz S, Stanslaski S, Oh Y, Rusheen AE, Klassen B, Miller K, Blaha CD, Bennet KE, Lee K. The development of an implantable deep brain stimulation device with simultaneous chronic electrophysiological recording and stimulation in humans. Biosens Bioelectron. 2021 Mar 15;176:112888. doi: 10.1016/j.bios.2020.112888. Epub 2020 Dec 15. PMID: 33395569; PMCID: PMC7953342.
- Jeong DU, Lim KM. Artificial neural network model for predicting changes in ion channel conductance based on cardiac action potential shapes generated via simulation. Sci Rep. 2021 Apr 9;11(1):7831. doi: 10.1038/s41598-021-87578-0. PMID: 33837240; PMCID: PMC8035260.
- Saha R, Wu K, Su D, Wang JP. Spin current nano-oscillator (SCNO) as a potential frequency-based, ultra-sensitive magnetic biosensor: a simulation study. Nanotechnology. 2020 Sep 11;31(37):375501. doi: 10.1088/1361-6528/ab9921. Epub 2020 Jun 3. PMID: 32492673.
- Saha R, Wu K, Bloom RP, Liang S, Tonini D, Wang JP. A review on magnetic and spintronic neurostimulation: challenges and prospects. Nanotechnology. 2022 Feb 10;33(18). doi: 10.1088/1361-6528/ac49be. PMID: 35013010.